Basal body movements orchestrate membrane organelle division and cell morphogenesis in Trypanosoma brucei.
Lacomble S, Vaughan S, Gadelha C, Morphew MK, Shaw MK, McIntosh JR, Gull K.
Journal of Cell Science (2010) 123:2884-2891.
PDF
In Focus
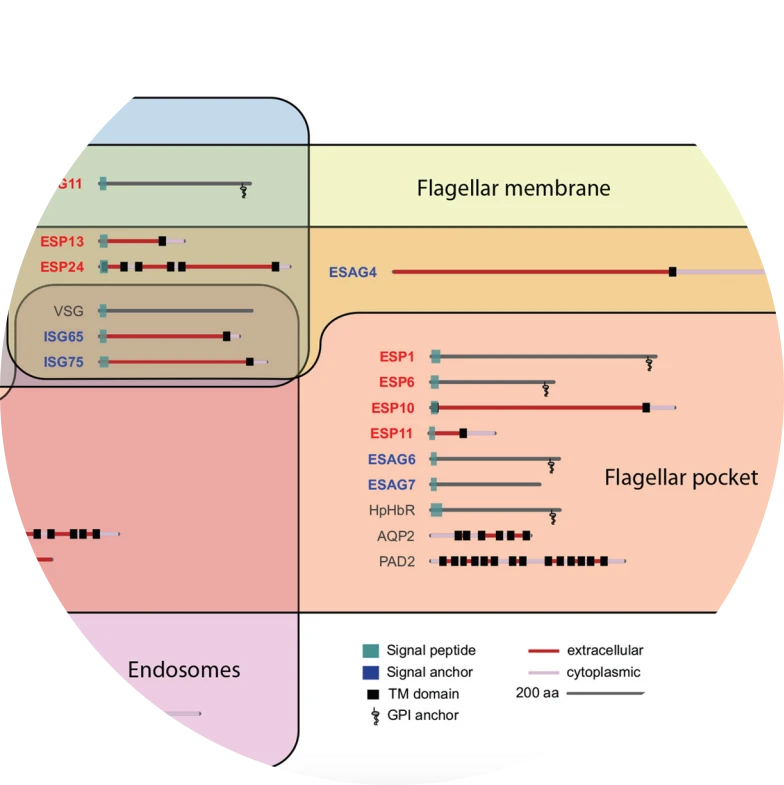
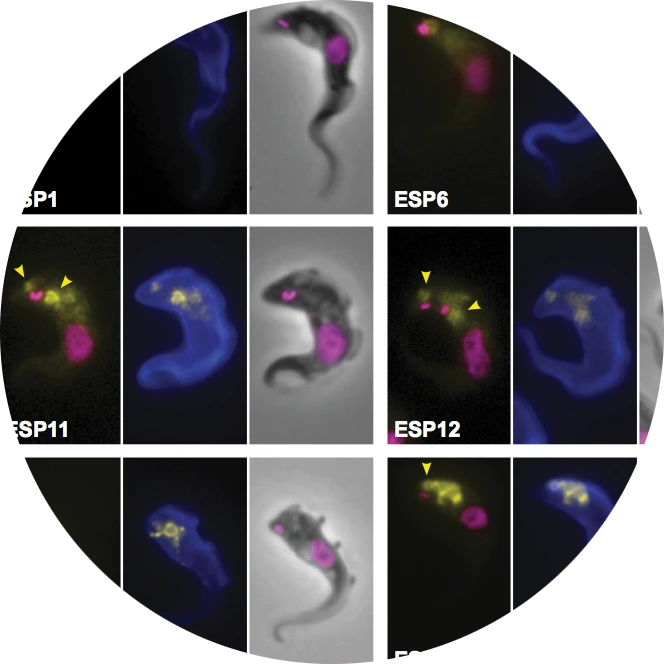
Membrane domains and flagellar pocket boundaries are influenced by the cytoskeleton in African trypanosomes.
Gadelha C, Rothery S, Morphew M, McIntosh JR, Severs NJ, Gull K.
Proceedings of the National Academy of Sciences of the U.S.A. (2009) 106:17425-17430.
PDF
Supplementary Figures
Three-dimensional cellular architecture of the flagellar pocket and associated cytoskeleton in trypanosomes revealed by electron microscope tomography.
Lacomble S, Vaughan S, Gadelha C, Morphew MK, Shaw MK, McIntosh JR, Gull K.
Journal of Cell Science (2009) 122:1081-1090.
PDF
In Focus
Flagellar and ciliary beating in trypanosome motility.
Gadelha C, Wickstead B, Gull K.
Cell Motility and the Cytoskeleton (2007) 64:629-643.
PDF
Electron microscopic tomography of the microtubule cytoskeleton and flagellar pocket in African trypanosomes.
Shaw M, Gadelha C, Lacomble S, Morphew M, O’Toole ET, McIntosh JR, Gull K.
Microscopy and Microanalysis (2007) 13:1310-1311.
PDF
Basal body and flagellum mutants reveal a rotational constraint of the central pair microtubules in the axonemes of trypanosomes.
Gadelha C, Wickstead B, McKean PG, Gull K.
Journal of Cell Science (2006) 119:2405-2413.
PDF
In Focus
Evidence for a sliding-resistance at the tip of the trypanosome flagellum.
Woolley D, Gadelha C, Gull K.
Cell Motility and the Cytoskeleton (2006) 63:741-746.
PDF
Cryptic paraflagellar rod in endosymbiont-containing kinetoplastid protozoa.
Gadelha C, Wickstead B, de Souza W, Gull K, Cunha-e-Silva N.
Eukaryotic Cell (2005) 4:516-525.
PDF
Improvement on the visualization of cytoskeletal structures of protozoan parasites using high-resolution field emission scanning electron microscopy (FESEM).
Sant’Anna C, Campanati L, Gadelha C, Lourenço D, Labati-Terra L, Bittencourt-Silvestre J, Benchimol M, Cunha-e-Silva NL, De Souza W.
Histochemistry and Cell Biology (2005) 124:87-95.
PDF
Relationships between the major kinetoplastid paraflagellar rod proteins: a consolidating nomenclature.
Gadelha C, LeBowitz JH, Manning J, Seebeck T, Gull K.
Molecular and Biochemical Parasitology (2004) 136:113-115.
PDF
Further studies on the structural analysis of the cuticle of Litomosoides chagasfilhoi (Nematoda: Filarioidea).
de Moraes Neto AHA, Lanfredi RM, Gadelha C, Cunha-e-Silva NL, Simão RA, Achete C, de Souza W.
Parasitology Research (2003) 89:397-406.
PDF